New insights into a useful killer protein
The cells that make up all complex life have evolved a variety of ways to undergo programmed cell death when necessary. There are many instances when one cell dying can benefit the entire organism – for example, a cell infected with a virus can sacrifice itself to hamper the spread of the virus to other healthy cells.
One programmed cell death pathway, known as necroptosis, is the focus of AINSE PGRA scholar Katherine Davies’ research. A virally infected cell that undergoes necroptosis will cause inflammation that, in turn, alerts the body’s immune system to the infection threat.
Necroptosis is mediated by a protein called MLKL (or Mixed Lineage Kinase domain-Like). When the inert MLKL protein is activated within a cell, multiple copies of the MLKL protein bind together, travel to the cell membrane and permeabilise this vital barrier. This results in the death of the cell.
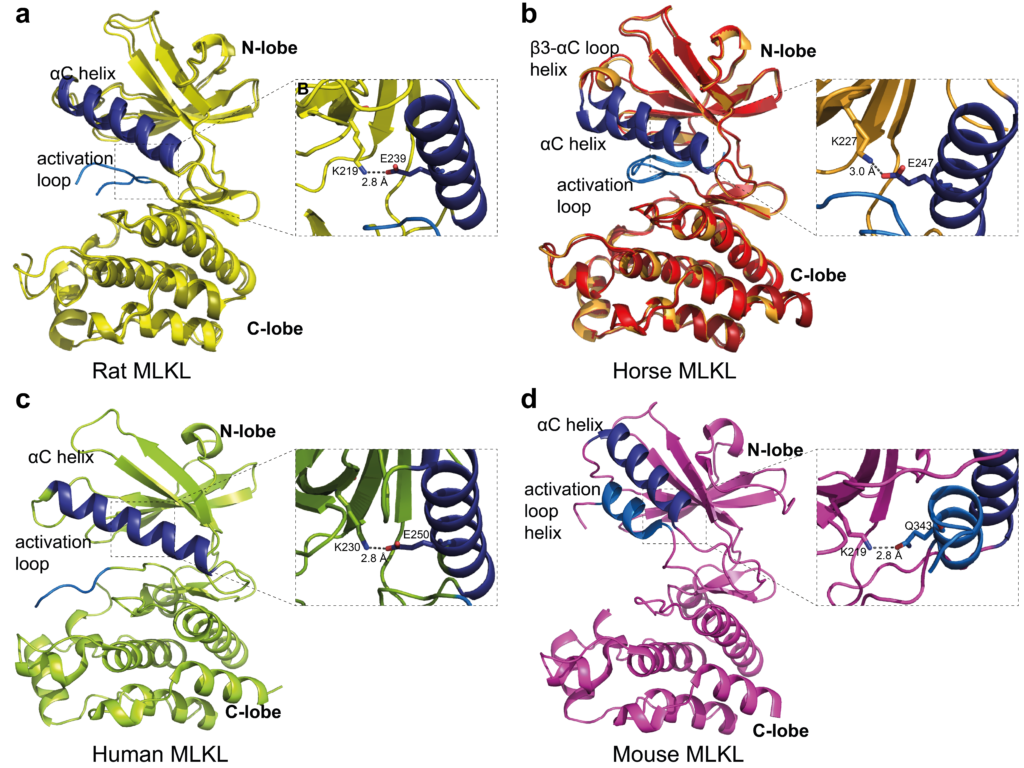
The exact process by which MLKL changes from an inert protein to a ‘killer protein’, however, is not yet well understood. This has important consequences for the development of new drugs that may target MLKL therapeutically. Usually, mice are used as the animal model of choice during the development of new medicines; however, multiple lines of evidence suggest that the mouse and human MLKL proteins differ slightly in their structure and activation mechanisms, meaning that the mouse system may not be a representative model of human necroptosis.
In order to gain further insight, Katherine – alongside her collaborators from the Walter and Eliza Hall Institute of Medical Research and the University of Melbourne – studied the structure of the MLKL proteins of rats and horses. The structure of these proteins was determined by X-ray crystallography, at the MX2 beamline of ANSTO’s Australian Synchrotron. This technique uses X-rays to interrogate protein crystals, in order to generate a diffraction pattern from which the structure of the protein can be determined. The Small Angle X-ray Scattering (SAXS) beamline was also used to investigate the protein structures in solution, to understand how multiple copies of the protein can bind together.
The investigation showed that rat MLKL protein’s structure, and behaviour in solution was more similar to human MLKL than the mouse protein, suggesting that rats may be a better model organism for further research into medicines that target MLKL for treating human disease.
To read more about the technical details of Katherine’s research, please see page 27 of the 2019 AINSE Annual Report.
Next Student Research Spotlight: James Hooper (Going back in time to understand the South American climate)
Previous Student Research Spotlight: Karthik Gopi (A new method for determining seafood provenance)
